Haussler-Salama lab and the UCSC Browser group publish the UCSC Repeat Browser
Mobile DNA | 31 March, 2020
Jason D. Fernandes, Armando Zamudio-Hurtado, W. James Kent, David Haussler, Sofie R. Salama, Maximilian Haeussler
Abstract
Background Nearly half the human genome consists of repeat elements, most of which are retrotransposons, and many of these sequences play important biological roles. However repeat elements pose several unique challenges to current bioinformatic analyses and visualization tools, as short repeat sequences can map to multiple genomic loci resulting in their misclassification and misinterpretation. In fact, sequence data mapping to repeat elements is often discarded from analysis pipelines. Therefore, there is a continued need for standardized tools and techniques to interpret genomic data of repeats.
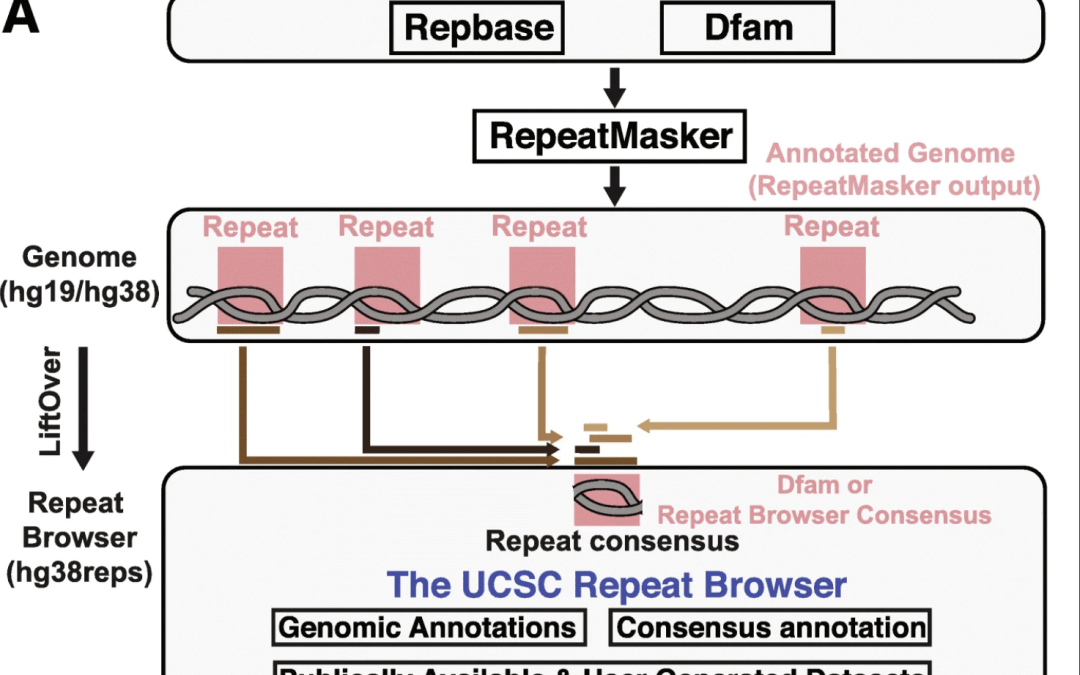
Results We present the UCSC Repeat Browser, which consists of a complete set of human repeat reference sequences derived from the gold standard repeat database RepeatMasker. The UCSC Repeat Browser contains mapped annotations from the human genome to these references and presents all of them as a comprehensive interface to facilitate work with repetitive elements. Furthermore, it provides processed tracks of multiple publicly available datasets of biological interest to the repeat community, including ChIP-SEQ datasets for KRAB Zinc Finger Proteins (KZNFs) – a family of proteins known to bind and repress certain classes of repeats. Here we show how the UCSC Repeat Browser in combination with these datasets, as well as RepeatMasker annotations in several non-human primates, can be used to trace the independent trajectories of species-specific evolutionary conflicts.
Conclusions The UCSC Repeat Browser allows easy and intuitive visualization of genomic data on consensus repeat elements, circumventing the problem of multi-mapping, in which sequencing reads of repeat elements map to multiple locations on the human genome. By developing a reference consensus, multiple datasets and annotation tracks can easily be overlaid to reveal complex evolutionary histories of repeats in a single interactive window. Specifically, we use this approach to retrace the history of several primate-specific LINE-1 families across apes and discover several species-specific routes of evolution that correlate with the emergence and binding of KZNFs.
