Pediatric Glioma
Pediatric Glioma
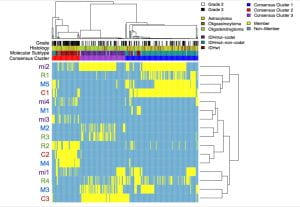
The Haussler-Salama lab became actively involved in cancer research in 2012 through their participation in the Cancer Genome Project (TCGA), where, along with the Josh Stuart lab, they played a key role in developing methods for identifying cancer driver pathways through integrative analysis of multiple genomic data types (i.e copy number variation, gene expression, DNA methylation). A major focus was gliomas were Salama was a member of the Glioma Analysis Working Group leadership. With then postdoctoral scholar, Olena Vaske, Haussler and Salama founded UCSC’s Treehouse Pediatric Cancer initiative, which applies these approaches to find treatment options for children whose tumors are refractory to standard treatments options or lack a standard of care.
Current lab projects are aimed at developing brain organoid-based preclinical models for pediatric gliomas using pluripotent stem cell lines engineered with known drivers of pediatric gliomas, primary tumor cell suspensions, and patient derived cell lines. We are testing the hypothesis that we can develop organoid-based models that are more realistic than standard 2-D cell lines, and more robust and higher throughput than animal-based models. This project is a close collaboration with the Vaske lab.
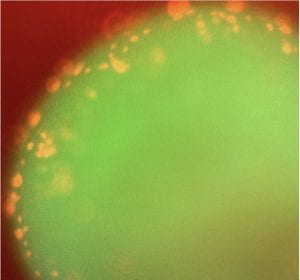
GFP expressing hindbrain organoid seven days after co-culture with TdTomato-expressing H3K27M Diffuse Intrinsic Pontine Glioma (DIPG) cell line (in red). Image by Quinn Brail.
Key Publications
Identification of a differentiation stall in epithelial mesenchymal transition in histone H3-mutant diffuse midline glioma