Haussler-Salama Lab undergraduate Taylor Real is awarded 2020 Dean’s Award
Congratulations to 2020 Dean’s Undergraduate Research Award winner Taylor Real, mentored by Dr. Sofie Salama of the Institute for the Biology of Stem Cells at UC Santa Cruz and the UC Santa Cruz Genomics Institute Scientific Director Dr. David Haussler.
Each year, the ten most excellent undergraduate theses or projects per academic division are selected for this award.
Taylor is majoring in molecular, cell, and developmental biology (MCDB) in the Division of Physical & Biological Sciences (UC Santa Cruz Science) and worked with MCDB graduate student Gary Mantalas on a method to better identify sequence variations in the human NOTCH2NL genes. These genes are involved in normal brain development and, if altered, potentially in diseases caused by abnormal brain development.
Optimizing Molecular Inversion Probe Assays to Characterize Human-Specific Neurodevelopmental Genes
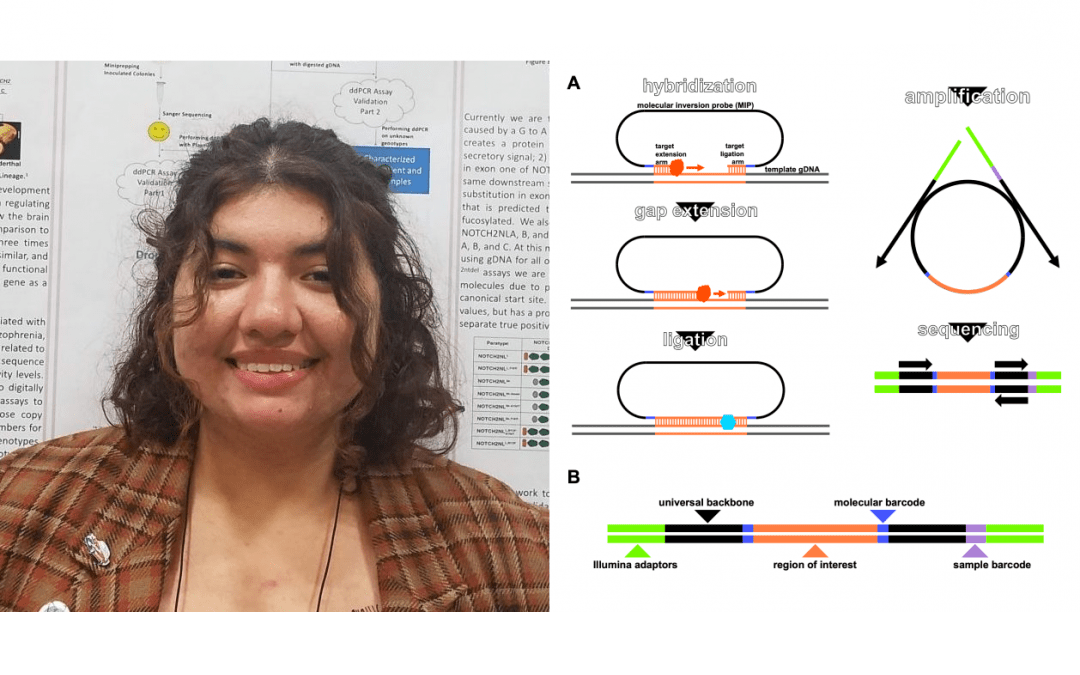
Copy number variation (CNV) of genes in the 2.3 Mb 1q21.1 duplication/deletion syndrome region of the human genome is highly correlated with neurodevelopmental disorders including microcephaly, macrocephaly, autism, and schizophrenia. NOTCH2NL is a human-specific gene with two paralogs within this region and we theorize that they contribute to the phenotypes of 1q21.1 duplication/deletion syndrome. NOTCH2NL is thought to regulate neural stem cell proliferation and thereby regulate the amount of neurogenesis in the developing brain. Population sequencing of NOTCH2NL genes revealed not only that paralogs A and B are breakpoints for the syndrome, but that there are small sequence variants (SSV) within different paralogs segregating in the population that we predict alter protein features, resulting in different activity levels. Our goal is to assess the total copy number of NOTCH2NL paralogs, as well as significant SSV in both neurotypical individuals and those with pathogenic 1q21.1 CNV events. To accomplish this we must first optimize Molecular Inversion Probes (MIPs), a PCR-based capture method. The MIPs protocol prepares products suitable for pooling a large number of samples for low cost, high-throughput sequencing. Molecular barcodes within the sequenced products are used to create ratios based on the numbers of reads corresponding to each allele detected within each MIP. Preliminary MIP experiments using gDNA with known genotypes have successfully captured NOTCH2NL variants. Characterizing NOTCH2NL allele profiles of those with typical and atypical neurodevelopment will help us understand the role of NOTCH2NL genes in the complex process of human brain development.
